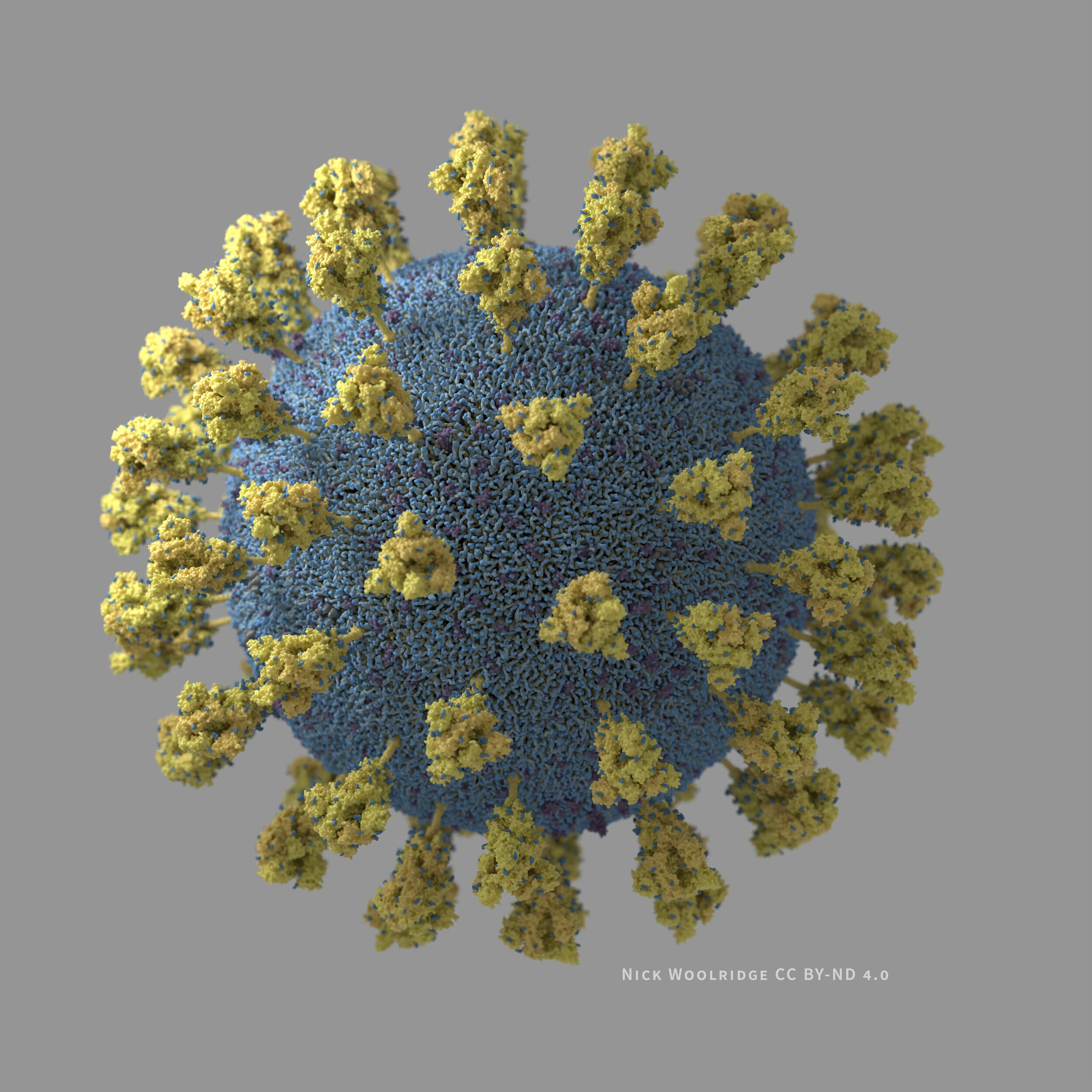
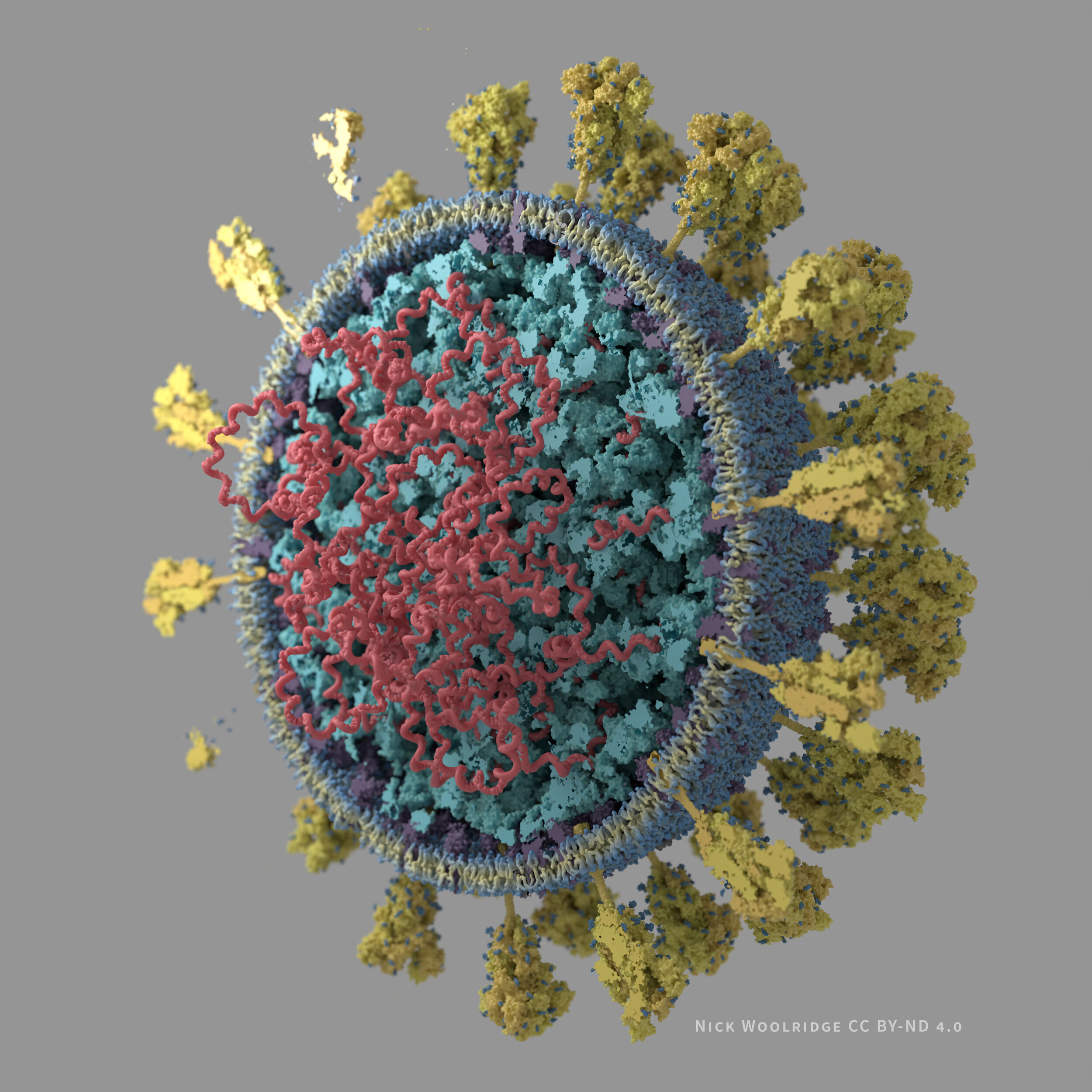
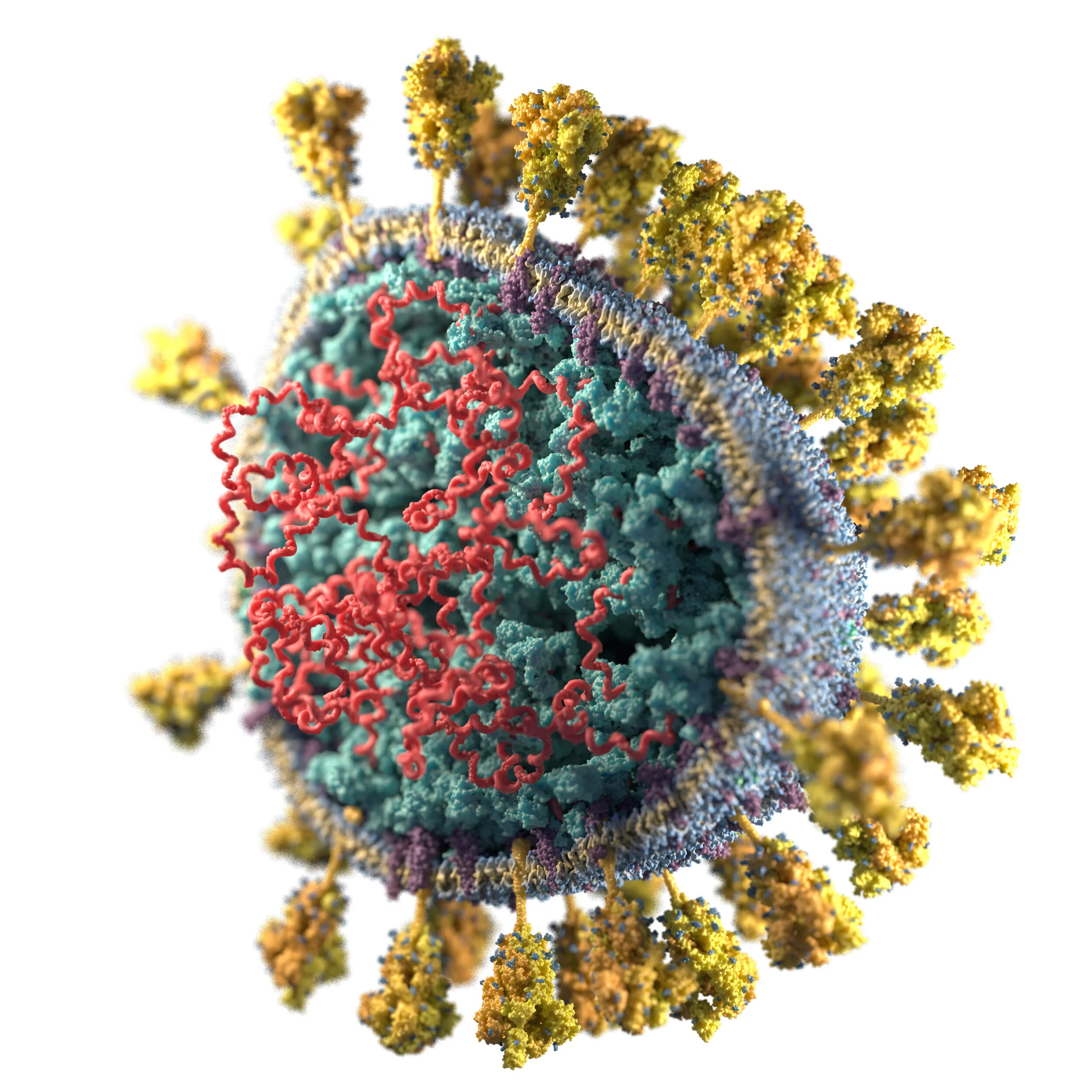
Images of SARS-CoV-2
I have created 3D visualizations/illustrations of the SARS-CoV-2 virus particle. The structures are based on the literature I was able to access as of late March 2020 (listed below).
Tech details: molecular structures were created with UCSF Chimera and ePMV. The models were assembled and animated in Cinema4D (making extensive use of its Mograph functionality), and the materials were developed and the images rendered in Arnold.
These images and animations are free for use under a Creative Commons Attribution-NoDerivatives 4.0 International License.
Please credit them to “Nick Woolridge / Biomedical Communications”.
Download animations:
Download 3000 x 3000 px images:
Cutaway view grey background .jpg
Whole view grey background .jpg
Cutaway pleomorphic white background .jpg
Cutaway pleomorphic transparent background .png
Limitations
These are illustrated visualizations, not the result of simulation or direct data acquisition
While most of the protein structures are derived from crystallographic/Cryo EM data, some (E-protein) are from the closely related SARS-CoV virus (not the SARS-CoV-2 virus causing the current outbreak). Some are computationally predicted from the genomic sequence (M-Protein and N-Protein).
Movement in the animated version is artistic license intended to convey the dynamic nature of the virus, and is not based on molecular dynamics simulation, temperature factors, or other data sources.
While I am relatively close to or within a first approximation of the raw numbers of most molecular players, the understanding of the structure of this virus will undoubtedly change as research proceeds.
The organization of the nucleoprotein region within the membrane is a rough approximation, and is currently “unphysical” (i.e. the molecules may intersect).
The lipid bilayer model is not the result of simulation, and currently only contains lipid molecules from a POPE membrane segment.
References/Data sources
A principal, but far from complete list:
Structural Proteins: PDB entries: 6VXX, 6VSB, 5X29 (from SARS-CoV), M-Protein as predicted by DeepMind, N-Protein as predicted by the Zhang Lab at UMich
The SARS coronavirus nucleocapsid protein – Forms and functions
https://www.sciencedirect.com/science/article/pii/S0166354213003781
Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1563832/
Electron microscopy studies of the coronavirus ribonucleoprotein complex.
https://link.springer.com/article/10.1007/s13238-016-0352-8